Translate this page into:
Overview of β-Lactam-Resistance Genes in Pandemic Multidrug-Resistant Acinetobacter baumannii: A Troublesome Pathogen in the Indian Intensive Care Unit

*Corresponding author: Dr. Manita Paneri Assistant Professor, Faculty of Health and Allied Sciences, Gurukashi University, Talwandi Sabo, Punjab, India. manitaprashant@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Paneri M, Sevta P, Yagnik VD, Gupta P, Bansal V, Sevta G, et al. Overview of β-Lactam-Resistance Genes in Pandemic Multidrug-Resistant Acinetobacter baumannii: A Troublesome Pathogen in the Indian Intensive Care Unit. Glob J Med Pharm Biomed Update. 2024;19:14. doi: 10.25259/GJMPBU_27_2024
Abstract
Objectives:
Acinetobacter baumannii is responsible for many infections in admitted patients, especially in the intensive care unit (ICU). Several risk factors may lead to an enhanced risk of A. baumannii colonization and infections. β-lactam antibiotics are frequently administered to treat Gram-positive and Gram-negative bacterial infections due to their minimum side effects, but the acquisition of β-lactamase genes has been the most challenging and troublesome situation and an imminent threat to the world as it increases mortality, medical expenses, and hospital stays. Hence, the present systematic review focused on the screening of β-lactam resistance genes that have been identified in the A. baumannii isolates’ genome and the nosocomial infections they cause in the Indian ICU.
Material and Methods:
This review has been done according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses guideline 2020. After screening, 317 genomes were included in this systematic review. We downloaded data from the bv-brc.org website on an Excel spreadsheet for statistical analysis. We presented categorical data in percentages (%) and in the form of a graph and pie chart.
Results:
Among the 317 isolates, pneumonia was caused by 189 strains (59.62%), bacteremia was caused by 109 strains (34.38%), respiratory infection by 12 isolates (3.79%), sepsis by 5 isolates (1.58%), and wound infection by 2 isolates (0.63%), which indicated that A. baumannii strains are highly involved in pneumonia followed by bacteremia. We did comparative genome analysis and found 26 β-lactamase genes; among them, the ADC2 gene was found to be in higher frequency (312) and was identified in 98.42% of A. baumannii isolates, followed by the OXA23 gene (303), which was found in 95.58% of isolates. The NDM-1 gene was identified in 181 (57.09%) isolates. OXA66 was found in 156 (49.21%) isolates. Our findings show a higher frequency of the ADC2 gene, followed by the OXA23 gene, in all these nosocomial infections. We have found that NDM-1, ADC2, OXA23, and OXA-66 genes coexisted in higher frequency in the A. baumannii isolates (137; 43.21%), followed by OXA23, OXA-66, and ADC2 (52; 16.40%).
Conclusion:
A. baumannii is a notorious pandemic pathogen, designated as a “priority of concern” by the World Health Organization. Our study indicates a high prevalence of the ADC2 gene, which gives resistance against the cephalosporin group and co-existence of β-lactamase genes (ADC2, OXA23, OXA66, and NDM-1) in various A. baumannii isolates’ genomes. This is a worrisome situation. Global molecular surveillance and the “One Health Concept” are crucial, as are research studies on plant extracts’ in vitro and in vivo efficacy against A. baumannii. Combating multidrug-resistant A. baumannii requires a multifaceted approach that involves infection control measures, antimicrobial stewardship, surveillance, education, research, and collaboration. Implementing these strategies and staying vigilant in the face of this resilient pathogen is essential to minimize its impact on health-care systems and public health.
Keywords
Acinetobacter
Antibiotic resistance
Beta-lactamase
Nosocomial infection
INTRODUCTION
Acinetobacter baumannii is a Gram-negative, catalase-positive, and oxidase-negative non-fermenting coccobacillus, and it belongs to the Moraxellaceae family. It is responsible for many infections in the admitted patients, especially in the intensive care unit (ICU). The infections caused by this notorious pathogen are “Hospital-acquired and ventilator-associated pneumonia, urinary tract infections, bacteremia, skin and soft-tissue infections, and meningitis.”[1-6] Its incidence in ICUs worldwide is up to 31%, and coinfection with SARS-CoV2 has been reported between 1 and 12.5%.[7-9]
Several risk factors may lead to an enhanced risk of A. baumannii colonization and infection. These include coexistent morbidities, prematurity in newborns, old age, major trauma, immunosuppressed conditions, major surgery, mechanical ventilation, presence of indwelling devices, extended hospital stay, and prior exposure to broad-spectrum antibiotics, for example, β-lactam antibiotics (Ampicillin, carbapenems, third-generation cephalosporins, and β-lactam + β-lactamase inhibitors), fluoroquinolones, aminoglycosides, etc. While attempting to overcome resistance, new broad-spectrum antimicrobials have fewer therapeutic alternatives.[10,11]
A. baumannii can develop antibiotic resistance through various mechanisms, including changing the antibiotic’s target area, managing how antibiotics penetrate its cell membrane, and enzyme-driven alterations of the antibiotics, making them ineffective. Beyond these typically genetically driven factors, A. baumannii can enhance its resistance using methods associated with its ability to cause disease. These include modifications in its outer cell protective layers, the uniqueness of the enzymes it produces, quorum sensing, and biofilms. It can achieve specific movement called twitching motility using hair-like structures (type IV pili) and has systems to obtain essential micronutrients. Furthermore, it employs specialized protein delivery methods and type II and VI secretion systems.[12,13]
While attempting to overcome resistance, new broad-spectrum antimicrobials have fewer therapeutic alternatives. Due to their limited side effects, β-lactam antibiotics are frequently administered to treat Gram-positive and Gram-negative bacterial infections. The β-lactam antibiotics exert their antibacterial activity by impeding the formation of the bacterial cell wall and have an enormously beneficial effect on managing life-threatening bacterial infections. However, they can be broken down by A. baumannii through several methods, including drug efflux pumps, drug target alterations, reduced membrane permeability, and the production of hydrolyzing enzymes. The acquisition of β-lactamase genes has been the most challenging and troublesome situation and an imminent threat to the world as it increases mortality, medical expenses, and hospital stays.[14]
Hence, the present systematic review focused on the screening of β-lactam resistance genes that have been identified in the genomes of A. baumannii isolates’ genome and nosocomial infections they cause in the Indian ICU.
MATERIAL AND METHODS
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline 2020 for this systematic review.[15] We have used the Bacterial and Viral Bioinformatic Resource Center (bv-brc.org) website for analysis of β-lactam resistance genes in A. baumannii isolates’ genome that have been collected and submitted from various parts of India. Their accession numbers are available on NCBI.[16] All the isolates are divided according to nosocomial infections they cause(bacteremia, pneumonia, other respiratory infections, sepsis, and wound infection) for genotypic analysis. For statistical analysis, we collected data on a Microsoft Excel spread-sheet. We presented categorical data in percentages (%) and in the form of graphs and pie charts.
RESULTS
Following the PRISMA guidelines 2020, we identified 14,474 genomes on the bv-brc.org website. By applying filters for “India,” “only human host,” “complete genome,” and “good quality,” we ultimately selected 317 A. baumannii isolates’ genomes for our β-lactam resistance gene analysis [Figure 1].
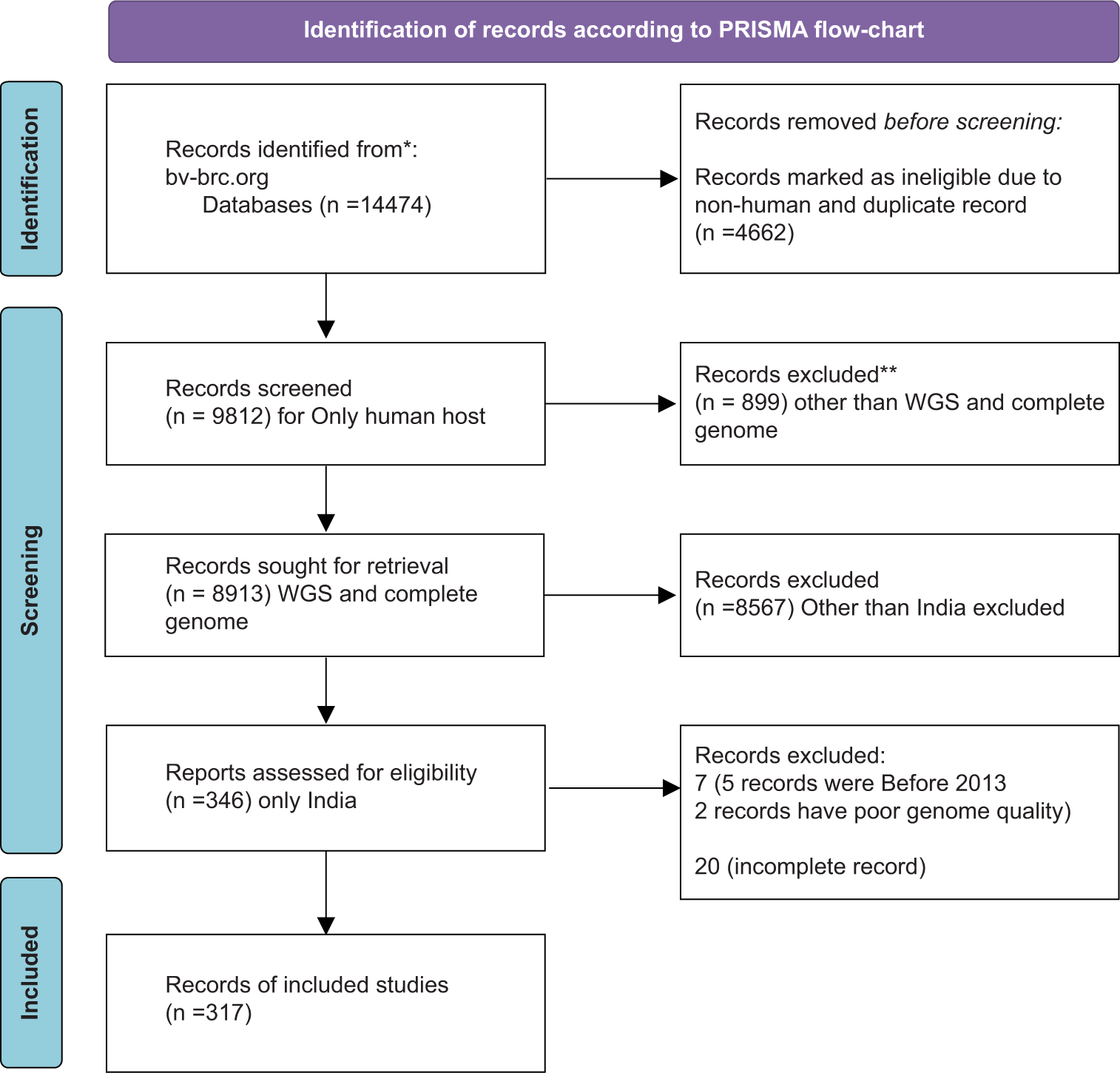
- Identification of records according to Preferred Reporting Items for Systematic Reviews and Meta-Analyses flow-chart 2020.
Genome sequencing was done using various platforms, such as Illumina HiSec, Ion torrent, PacBio, and Oxford Nanopore MiniIon, and various assembly methods were used in these Bio projects such as SPAdes v. 5, SPAdes 3.14.1, Canu v.v 1.6, Canu v.v 1.7, UniCycler v.0.4.6, and UniCycler v.0.4.8. Typing was done through multilocus sequence typing. Data were then downloaded from bv-brc.org onto an Excel spreadsheet for further analysis. The mean coding sequence was 3900. The average genome size was found to be 3,995,080 bp. The maximum genome size was found in A. baumannii SP 1917, which harbors the AmpC gene and is involved in pneumonia whereas A. baumannii strain SP25 had a minimum genome size of 3,620,806 bp. A. baumannii strain VB958 contained the maximum number of transfer RNA (tRNA) (76) whereas the minimum number (25) of tRNA was found in A. baumannii BA22708. The average G + C% content of all A. baumannii isolates was 38.54%.
Descriptive statistics analysis
We divided the total 317 A. baumannii isolates into five groups according to the infections they caused, which were bacteremia, pneumonia, other respiratory infections, sepsis, and wound infection. Among 317 isolates, pneumonia was caused by 189 strains (59.62%), bacteremia by 109 strains (34.38%), respiratory infection by 12 isolates (3.79%), sepsis by 5 isolates (1.58%), and wound infection by 2 isolates (0.63%). This indicated that A. baumannii strains are highly involved in pneumonia, followed by bacteremia [Table 1 and Figure 2].
| Variable | Level | Total no. of isolates | Count | Proportion | P-value |
|---|---|---|---|---|---|
| Host health | Bacteremia | 317 | 109 | 0.34 | <0.001 |
| Pneumonia | 317 | 189 | 0.596 | <0.001 | |
| Other respiratory infection | 317 | 12 | 0.038 | <0.001 | |
| Sepsis | 317 | 5 | 0.016 | <0.001 | |
| Wound infection | 317 | 2 | 0.006 | <0.001 |
n=317 Acinetobacter baumannii isolates, Confidence level 95%, Bold value indicates higher value of Proportion
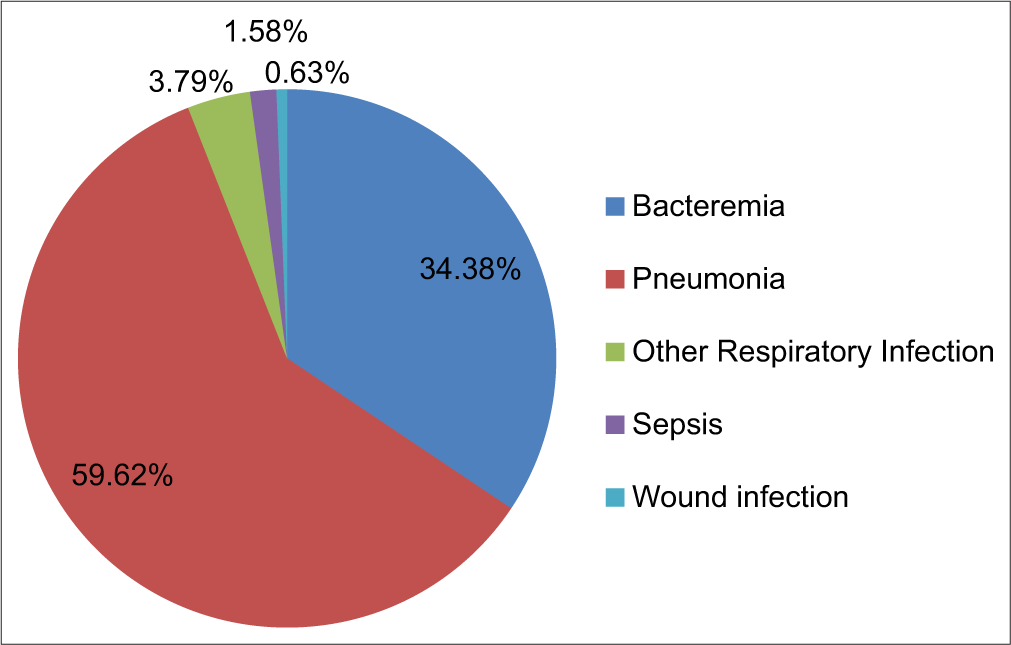
- Graphical representation of host health and Acinetobacter baumannii isolates (%).
Effect of β-lactam resistance genes in host’s health
We identified a total of 26 β-lactam resistance genes in the various A. baumannii isolates’ genomes, which are given in Tables 2 and 3, Figure 3. Mostly, these genes are found to be involved in pneumonia (mean score 24.19 ± 53.83 standard deviation [SD]) followed by bacteremia (mean score 15.57 ± 32.17 SD).
| Gene group | Total no. of isolates |
|---|---|
| OXA23, OXA144, NDM1, ADC2 | 2 |
| OXA208, ADC2 | 1 |
| OXA23, OXA104, ADC2 | 3 |
| OXA23, OXA104, PER7, ADC2 | 4 |
| OXA23, OXA104, PER7, NDM1, ADC2 | 11 |
| OXA23, OXA144, PER7, CARB3, ADC2 | 1 |
| OXA23, OXA180, OXAADC2 | 1 |
| OXA23, OXA27, OXA49, NDM1, ADC2 | 1 |
| OXA23, OXA27, OXA68, PER7 | 1 |
| OXA23, OXA371, ADC2 | 1 |
| OXA23, OXA371, NDM1, ADC2 | 3 |
| OXA23, OXA49, OXA66, NDM1, ADC2 | 1 |
| OXA23, OXA49, OXA64, ADC2, PER7 | 1 |
| OXA23, OXA49, OXA66, ADC2 | 1 |
| OXA23, OXA58, OXA68, PER7, ADC2 | 1 |
| OXA23, OXA64, ADC2 | 4 |
| OXA23, OXA64, PER7, ADC2 | 8 |
| OXA23, OXA66, OXA98, NDM1, ADC2 | 2 |
| OXA23, OXA66, ADC2 | 52 |
| OXA23, OXA66, ADC2, PER7 | 16 |
| OXA23, OXA66, ADC2, TEM1 | 25 |
| OXA23, OXA66, NDM1, ADC2 | 137 |
| OXA23, OXA66, NDM1, TEM1 | 1 |
| OXA23, OXA68, OXA420, ADC2, CARB1, PER7 | 1 |
| OXA23, OXA68, ADC2 | 1 |
| OXA23, OXA68, NDM1, ADC2 | 4 |
| OXA23, OXA68, PER7, ADC2 | 3 |
| OXA23, OXA69, NDM1, ADC2 | 13 |
| OXA23, OXA70, ADC2 | 1 |
| OXA23, OXA98, ADC2, NDM1 | 2 |
| OXA33, ADC2 | 1 |
| OXA33, AmpC | 1 |
| OXA371, NDM1, ADC2 | 1 |
| OXA58, OXA64, CARB1, NDM1, ADC2 | 2 |
| OXA67, ADC2 | 2 |
| OXA68, OXA23, NDM1, ADC2 | 1 |
| OXA68, PER7, ADC2 | 2 |
| OXA98, ADC2 | 2 |
| AmpC | 1 |
| PER1, AmpC | 1 |
| Genes | Bacteremia | Pneumonia | Other respiratory infection | Sepsis | Wound infection | Total |
|---|---|---|---|---|---|---|
| ADC2 | 107 | 187 | 12 | 4 | 2 | 312 |
| NDM-1 | 51 | 117 | 10 | 2 | 1 | 181 |
| TEM-1 | 13 | 12 | 1 | 26 | ||
| AmpC | 2 | 1 | 3 | |||
| PER1 | 1 | 1 | ||||
| PER7 | 18 | 19 | 10 | 3 | 50 | |
| CARB1 | 2 | 1 | 3 | |||
| CARB3 | 1 | 1 | ||||
| OXA23 | 100 | 184 | 12 | 5 | 2 | 303 |
| OXA27 | 2 | 2 | ||||
| OXA33 | 2 | 2 | ||||
| OXA49 | 1 | 1 | 2 | 4 | ||
| OXA58 | 3 | 3 | ||||
| OXA64 | 6 | 7 | 1 | 14 | ||
| OXA66 | 87 | 66 | 1 | 1 | 1 | 156 |
| OXA67 | 2 | 2 | ||||
| OXA68 | 3 | 6 | 1 | 1 | 11 | |
| OXA69 | 1 | 11 | 12 | |||
| OXA70 | 1 | 1 | ||||
| OXA98 | 2 | 4 | 6 | |||
| OXA104 | 1 | 7 | 9 | 1 | 18 | |
| OXA144 | 1 | 1 | 1 | 3 | ||
| OXA180 | 1 | 1 | ||||
| OXA208 | 1 | 1 | ||||
| OXA371 | 2 | 3 | 5 | |||
| OXA420 | 1 | 1 |
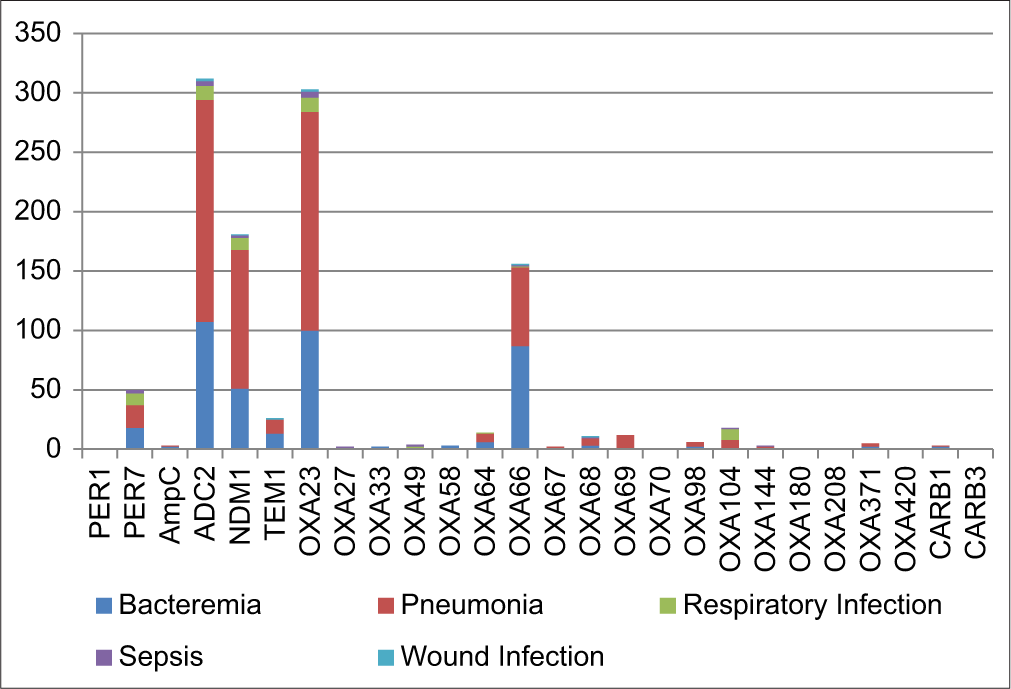
- β-lactam resistant genes that have been identified in various Acinetobacter baumannii isolates’ genome.
Among 26 β-lactamase genes, the ADC2 gene was found to be more frequent (312) and was identified in 98.42% of A. baumannii isolates, followed by OXA23 gene (303), which was found in 95.58% isolates. NDM-1 gene was identified in 181 (57.09%) isolates. OXA66 was found in 156 (49.21%) isolates.
We did a comparative genome analysis and found that among all the other β-lactam resistance genes, the ADC2 gene, followed by the OXA23 gene, was highly involved in pneumonia, followed by bacteremia among other infections. Our findings show a higher frequency of the ADC2 gene, followed by the OXA23 gene, in all nosocomial infections. We found that NDM-1, ADC2, OXA23, and OXA-66 genes coexisted in higher frequency in the A. baumannii isolates (137; 43.21%), followed by OXA23, OXA-66, and ADC2 gene (52; 16.40%).
In 95 (69.34%) out of 137 A. baumannii isolates, the OXA23, OXA-66, NDM-1, and ADC2 genes are jointly involved in pneumonia, and in 41 isolates (30%), they are found to be associated with bacteremia, and only one isolate caused other respiratory infection. In 31 (59.61%) out of 52 A. baumannii strains, OXA23, OXA-66, and ADC2 coexisting genes were involved in pneumonia, and 21 strains (40.38%) caused bacteremia.
DISCUSSION
A. baumannii, a notorious pathogen known for causing most hospital-acquired infections, is becoming more menacing. Known for its unmatchable virulence and pathogenicity, it has become a consistent and prominent concern for intensivists and infectious diseases specialists.[17,18] A. baumannii has acquired the capability to cause untreatable infections gradually through various mechanisms such as efflux pumps, reduction or inactivation of expression of porins, modification in expression or synthesis of new penicillin-binding proteins and presence of β-lactamases.[11,14,19] Among these, the presence of β-lactamases is the most prominent and p seriously threatens mankind. Based on sequence homology, β-lactamases are grouped into molecular classes A, B, C, and D.[20,21] All four classes of β-lactamases have been identified in A. baumannii.[20]
Class A beta-lactamases are classified as serine beta-lactamases due to the presence of Serine in the enzyme’s active site. The most prevalent Class A beta-lactamases are TEM and SHV. Our study found TEM-1 in 26 isolates, with 13 involved in bacteremia, 12 in pneumonia, and 1 in wound infection. A. baumannii often possesses plasmids that encode extended-spectrum beta-lactamases, along with resistance to other antimicrobials (e.g., narrow-spectrum cephalosporin), including aminoglycosides and fluoroquinolones. Shali et al., in 2022, found that all A. baumannii isolates collected from burn patients exhibited TEM-1, giving resistance against penicillin and cephalosporin classes.[22]
C lass B Metallo β-lactamases differ from other classes by being Metallo-lactamases as opposed to other classes, which are serine β-lactamases. As a result, these enzymes require metallic ions to act, namely Zn2+. They are further divided into B1, B2, and B3, depending on the number of Zn2+ ions being utilized. The most notorious enzyme in this class is NDM-1 (New Delhi Metallo β lactamases, first isolated from a urine sample in New Delhi).
In our comparative genome analysis, we identified the NDM-1 gene in 181(57.09%) isolates, whereas Sharma et al., 2023 found that out of 317 isolates, 189 (59.62%) were harboring the NDM-1 gene.[23] The strains producing other variants, such as NDM-2, NDM-5, NDM-6, or NDM-42, have been sporadically reported.[24-28]
Various other enzymes, such as NDM (over 24 subtypes), IMP, GMP, and VIM, have been identified in this class. The action of Tn125 likely facilitates the acquisition of the NDM-1 gene. The presence of integron 1 is the main driving force behind bacteria acquiring this enzyme, which also confers the property of other antibiotic resistance, namely, aminoglycosides. Since integron 1 is located over plasmids, which are easily transferred horizontally, widespread dissemination is their hallmark. This has been seen in other bacteria as well. Even with the chelation of heavy metal ions, NDM does not lose its potency as it is anchored to the outer membrane protein, which prevents its destabilization. blaNDM-1 is typically linked to several genetic determinants that indicate resistance to a wide range of antibiotics, leaving only last-resort antibiotics – which are usually employed in combination therapies – as a viable treatment option.[29]
Class C β-lactamases comprise mainly Acinetobacter derived cephalosporinases (ADCs), which are intrinsic to this bacterium. The presence of ISAba1 causes overexpression of AmpC, which belongs to the same family of Class C β-lactamases.[30] They usually confer resistance to β-lactam inhibitors but are inhibited by cloxacillin or boronic acid. Rao et al. (2020) found out the ADC gene in all A. baumannii isolates, whereas in our systematic review, we identified the ADC2 gene in 98.42% of isolates.[31] No further research work has been done on the ADC2 gene in India, yet according to our comparative analysis, it has been found in almost all A. baumannii isolates.
Class D β-lactamases, also called oxacillinases (OXAs) as they hydrolyze oxacillin better than benzylpenicillin, are usually plasmid-mediated. Since the 1980s, more than 400 types of OXAs have been identified, such as OXA-23, OXA-24/40, OXA-58, OXA-143, and OXA-235.[32] The bla gene, almost in all its subtypes, is preceded by an IS element, ISAba1 or ISAba4, which leads to its overexpression, hence is the prime reason for its dissemination throughout the globe.
We have depicted in our previous article that OXA23 (314; 92.62%) followed by OXA66 (241; 71.09%) has a higher frequency in A. baumannii isolates,[10] and more research work is required on this c lass D enzymes due to its weak hydrolysis activity. Researchers neglected this class and only focused their work on other virulent carbapenemase. In our present study, we identified the OXA23 gene in 95.58% isolates’ genome. Kumar et al. (2019)found a high prevalence of OXA23 genes (97.7%) in A. baumannii isolates.[28]
CONCLUSION
A. baumannii is a notorious pandemic pathogen designated as a “priority of concern” by the World Health Organization. Our study indicates a high prevalence of the ADC2 gene, which gives resistance against the cephalosporin group and co-existence of β-lactamase genes (ADC2, OXA23, OXA66, and NDM-1) in various A. baumannii isolates’ genomes, which is a problematic situation. Global molecular surveillance and the “One Health Concept” are crucial, as is research on plant extracts in vitro and in vivo efficacy against A. baumannii. Combating multidrug-resistant A. baumannii requires a multifaceted approach that involves infection control measures, antimicrobial stewardship, surveillance, education, research, and collaboration. Implementing these strategies and staying vigilant in the face of this resilient pathogen is essential to minimize its impact on healthcare systems and public health.
Author contribution
All the authors contributed equally.
Ethical approval
Institutional Review Board approval is not required.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
Nil.
References
- Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J Infect Dis. 2008;197:1079-81.
- [CrossRef] [PubMed] [Google Scholar]
- Acinetobacter baumannii an evolving and cunning opponent. Front Microbiol. 2024;22:15:1332108.
- [CrossRef] [PubMed] [Google Scholar]
- The Global Prevalence of Multidrug-resistance among Acinetobacter baumannii Causing Hospital-acquired and Ventilator-associated Pneumonia and Its Associated Mortality: A Systematic Review and Meta-analysis. J Infect. 2019;79:593-600.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug Resistant and Extensively Drug-resistant Acinetobacter baumannii Hospital Infection Associated with High Mortality: A Retrospective Study in the Pediatric Intensive Care Unit. BMC Infect Dis. 2020;20:597.
- [CrossRef] [PubMed] [Google Scholar]
- The Association between Acinetobacter baumannii Infections and the COVID-19 Pandemic in an Intensive Care Unit. Sci Rep. 2022;12:20808.
- [CrossRef] [PubMed] [Google Scholar]
- Evaluation of Acinetobacter baumannii Pneumonia among Critically Ill Patients in a Tertiary Care Hospital in Saudi Arabia. Heliyon. 2020;6:e03976.
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiological and Clinical Characteristics of 99 Cases of 2019 Novel Coronavirus Pneumonia in Wuhan, China: A Descriptive Study. Lancet. 2020;395:507-13.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug-resistant Infections and Outcome of Critically Ill Patients with Coronavirus Disease 2019: A Single Center Experience. Microb Drug Resist. 2021;27:1167-75.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug-resistant Acinetobacter baumannii Infections: Looming Threat in the Indian Clinical Setting. Expert Rev Anti Infect Ther. 2022;20:721-32.
- [CrossRef] [PubMed] [Google Scholar]
- Burden of Carbapenem Resistant Acinetobacter baumannii Harbouring BLA OXA Genes in the Indian Intensive Care Unit. Glob J Med Pharm Biomed Update. 2023;18:12.
- [CrossRef] [Google Scholar]
- Insights into Acinetobacter baumannii A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics (Basel). 2020;9:119.
- [CrossRef] [PubMed] [Google Scholar]
- Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens. 2021;10:373.
- [CrossRef] [PubMed] [Google Scholar]
- Efficacy of Plant Extracts against Carbapenem Resistant Acinetobacter baumannii A Notorious Pathogen in the Intensive Care Unit. Glob J Med Pharm Biomed Update. 2023;18:24.
- [CrossRef] [Google Scholar]
- Overview of Antimicrobial Resistance: An Emerging Silent Pandemic. Glob J Med Pharm Biomed Update. 2023;18:11.
- [CrossRef] [Google Scholar]
- The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. Int J Surg. 2021;88:105906.
- [CrossRef] [PubMed] [Google Scholar]
- Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A Resource Combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51:D678-89.
- [CrossRef] [PubMed] [Google Scholar]
- Biology of Acinetobacter baumannii Pathogenesis, Antibiotic Resistance Mechanisms, and Prospective Treatment Options. Front Cell Infect Microbiol. 2017;7:55.
- [CrossRef] [PubMed] [Google Scholar]
- adeABC Efflux Gene in Acinetobacter baumannii. New Microbes New Infect. 2019;30:100549.
- [CrossRef] [PubMed] [Google Scholar]
- The Structure of Beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321-31.
- [CrossRef] [PubMed] [Google Scholar]
- β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J Mol Biol. 2019;431:3472-500.
- [CrossRef] [PubMed] [Google Scholar]
- Structural Basis for Carbapenem-hydrolyzing Mechanisms of Carbapenemases Conferring Antibiotic Resistance. Int J Mol Sci. 2015;16:9654-92.
- [CrossRef] [PubMed] [Google Scholar]
- Dissemination and Genetic Relatedness of Multidrug-Resistant and Extensively Drug-Resistant Acinetobacter baumannii Isolates from a Burn Hospital in Iraq. Can J Infect Dis Med Microbiol. 2022;2022:8243192.
- [CrossRef] [PubMed] [Google Scholar]
- Susceptibility Profile of blaOXA-23 and Metallo-β-lactamases Co-harbouring Isolates of Carbapenem-resistant Acinetobacter baumannii (CRAB) against Standard Drugs and Combinations. Front Cell Infect Microbiol. 2022;12:1068840.
- [CrossRef] [PubMed] [Google Scholar]
- Beta-lactamase Database (BLDB)-Structure and Function. J Enzyme Inhib Med Chem. 2017;32:917-9.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic Characterization of an NDM-9-Producing Acinetobacter baumannii Clinical Isolate and Role of Glu152Lys Substitution in the Enhanced Cefiderocol Hydrolysis of NDM-9. Front Microbiol. 2023;14:1253160.
- [CrossRef] [PubMed] [Google Scholar]
- Clonal Diversity of Acinetobacter Clinical Isolates Producing NDMtype Carbapenemase in Cuba, 2013-19. IJID Reg. 2022;5:93-6.
- [CrossRef] [PubMed] [Google Scholar]
- Outbreak of Efficiently Transferred Carbapenem-Resistant blaNDM-Producing Gram-Negative Bacilli Isolated from Neonatal Intensive Care Unit of an Indian Hospital. Microb Drug Resist. 2020;26:284-9.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular Epidemiology of Carbapenem-resistant Acinetobacter baumannii Isolates Reveals the Emergence of blaOXA-23 and blaNDM-1 Encoding International Clones in India. Infect Genet Evol. 2019;75:103986.
- [CrossRef] [PubMed] [Google Scholar]
- Genomic Characterization of Mobile Genetic Elements Associated With Carbapenem Resistance of Acinetobacter baumannii From India. Front Microbiol. 2022;13:869653.
- [CrossRef] [PubMed] [Google Scholar]
- Distribution of Carbapenem-resistant Acinetobacter baumannii with blaADC-30 and Induction of ADC-30 in Response to Beta-lactam Antibiotics. Res Microbiol. 2020;171:128-33.
- [CrossRef] [PubMed] [Google Scholar]
- Detection of Antimicrobial Resistance Genes Associated with Carbapenem Resistance from the Whole-Genome Sequence of Acinetobacter baumannii Isolates from Malaysia. Can J Infect Dis Med Microbiol. 2020;2020:5021064.
- [CrossRef] [PubMed] [Google Scholar]
- Origin of OXA-23 Variant OXA-239 from a Recently Emerged Lineage of Acinetobacter baumannii International Clone V. mSphere. 2020;5:e00801-19.
- [CrossRef] [PubMed] [Google Scholar]







