Translate this page into:
Burden of Carbapenem Resistant Acinetobacter baumannii Harboring blaOXA Genes in the Indian Intensive Care Unit

*Corresponding author: Manita Paneri, Department of Medical Microbiology, Centre for Interdisciplinary Biomedical Research, Adesh University, Bathinda, Punjab, India. manitaprashant@gmail.com
-
Received: ,
Accepted: ,
How to cite this article: Paneri M, Sevta P, Yagnik VD. Burden of carbapenem-resistant Acinetobacter baumannii harboring blaOXA genes in the Indian intensive care unit. Glob J Med Pharm Biomed Update 2023;18:12.
Abstract
Objectives:
The World Health Organization (WHO) mentioned Acinetobacter baumannii as a “priority of concern” in 2017. Acinetobacter baumannii generally infects immunocompromised patients and causes various nosocomial infections in the intensive care unit (ICU) such as bacteremia, meningitis, ventilator-associated pneumonia, other respiratory infections, and surgical site infections. As oxacillinase has weak hydrolysis activity, more work was needed on this class-D beta-lactamase. Hence, the current Systematic review focuses on the A. baumannii’s oxacillinase (Class-D beta-lactamases) enzyme and its variants collected during 2013–2020 in India for complete genome sequencing.
Method:
This Systematic review has been done according to PRISMA guideline 2020. We have used the Bacterial and Viral Bioinformatic Resource Centre (bv-brc.org) system for comparative genome analysis. The protein Basic Local Alignment Search Tool (BLAST) was used to identify similarities between sequences, in which BLOSUM62 was used as a scoring matrix. Clustal-W was used for multiple sequence alignment. A phylogenetic tree of the blaOXA gene family has been constructed using MEGA version 11.
Result:
In India during 2013–2020, for genome sequencing of A. baumannii, the highest number of samples was collected from blood (36%), following the ETA (30%). The average G+C % content was 38.95%. Among the 339 A. baumannii isolates, a maximum of 189 (55.75%) strains caused pneumonia, whereas 113 (33.33%) strains were involved in bacteremia. Carbapenems seemed effective, but resistance against them was higher. Among all A. baumannii genomes, bla-OXA-23 had the highest frequency (314; 92.62%), followed by bla-OXA-66 (241; 71.09%) in India.
Conclusion:
Our findings indicated that a high percentage of A. baumannii strains that produce oxacillinases exist in India, emphasizing the necessity for indigenous molecular surveillance to assist effective management and preventative initiatives. Comparative genomics and next-generation sequencing will offer tremendous potential for tracking and regulating the spread of this dangerous bacterium.
Keywords
Acinetobacter baumannii
Carbapenem
oxacillinase
Antibiotic-resistant
Antimicrobials
INTRODUCTION
Acinetobacter baumannii is an opportunistic and non-fermenting gram-negative bacillus that belongs to the Moraxellaceae family and inhabits hospital environments. The World Health Organization (WHO) mentioned it as a “critical priority of concern” in 2017.[1] A. baumannii generally infects immunocompromised patients and causes various nosocomial infections in the intensive care unit (ICU) such as bacteremia, meningitis, ventilator-associated pneumonia, other respiratory infections, and surgical site infections.[2]
Carbapenems are beta-lactam antibiotics administered to critically ill patients and considered as a last resort to treat infections caused by gram-negative bacteria, especially multidrug-resistant pathogens such as A. baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa etc.[3] Due to the extensive use of these antibiotics in the ICU as an empirical treatment, the bacterial genome was under selection pressure. Through intrinsic and extrinsic resistance, these pathogens have developed various defensive mechanisms such as overexpression of efflux pump, loss of porin channel, modifying, or hydrolyzing antibiotics, etc. to combat with antimicrobials, and even through horizontal gene transfer and transposons, cross-species transfer of these resistant genes among different gram-negative bacteria is also possible.[4] As a result, these therapeutics are now less effective than before. There are no new medicines on the market to treat infection caused by multidrug resistance (MDR) and extensive drug resistance (XDR) pathogens.
In the last decade, research was mainly focused on virulent resistance genes such as NDM-1, TEM-1, and KPC, etc., and after the COVID outbreak, review studies were flooded with COVID-19 articles. As oxacillinase has weak hydrolysis activity, more work was needed on this class-D beta-lactamases. There is no relevant comparative data about oxacillinase-encoding genes (blaOXA) that provide resistance to A. baumannii against carbapenems in the Indian scenario. Hence, the current Systematic review focuses on the A. baumannii’s oxacillinase (Class-D beta-lactamases) enzyme and its variants.
METHODOLOGY
This Systematic review has been done according to PRISMA guideline 2020.[5] We have used the Bacterial and Viral Bioinformatic Resource Centre (bv-brc.org) system for comparative genome analysis.[6,7] Comparative charts have been prepared for the year-wise collection of isolates, sample sources, and various bla-OXA genes that have been found in the A. baumannii genome.
For further analysis, FASTA sequences for all the oxacillinase-encoding genes (bla OXA) were retrieved from National Centre for Biotechnology Information (NCBI). Protein Basic Local Alignment Search Tool (BLAST) was used to identify similarities between sequences, in which for the scoring the BLOSUM62 matrix was used.[8] Clustal-W was used for multiple sequence alignment. A phylogenetic tree of the bla-OXA gene family has been constructed using MEGA version 11 software with bootstrap values corresponding to 1000 replications. The evolutionary history was inferred using the neighbor-joining method.[9-11]
RESULTS
We have found 14,474 genomes of A. baumannii as of December 29, 2022, and by following PRISMA guidelines, we have finally selected 339 genomes of A. baumannii. We have included only those A. baumannii isolates found in human hosts from India during 2013–2020 for comparative genome analysis. Isolates from non-human hosts, isolates from countries other than India, low-quality genomes, and records submitted before 2013 were all excluded from the study. A filter has been applied to select the desired genome using the following words: “complete,” “WGS,” “good quality,” and “India.” The gene accession number of all isolates are available on the NCBI website. The mean coding sequence (CDS) was 3909. The Illumina HiSeq sequencing platform was used for the genome sequencing of most of the A. baumannii isolates [Figure 1].
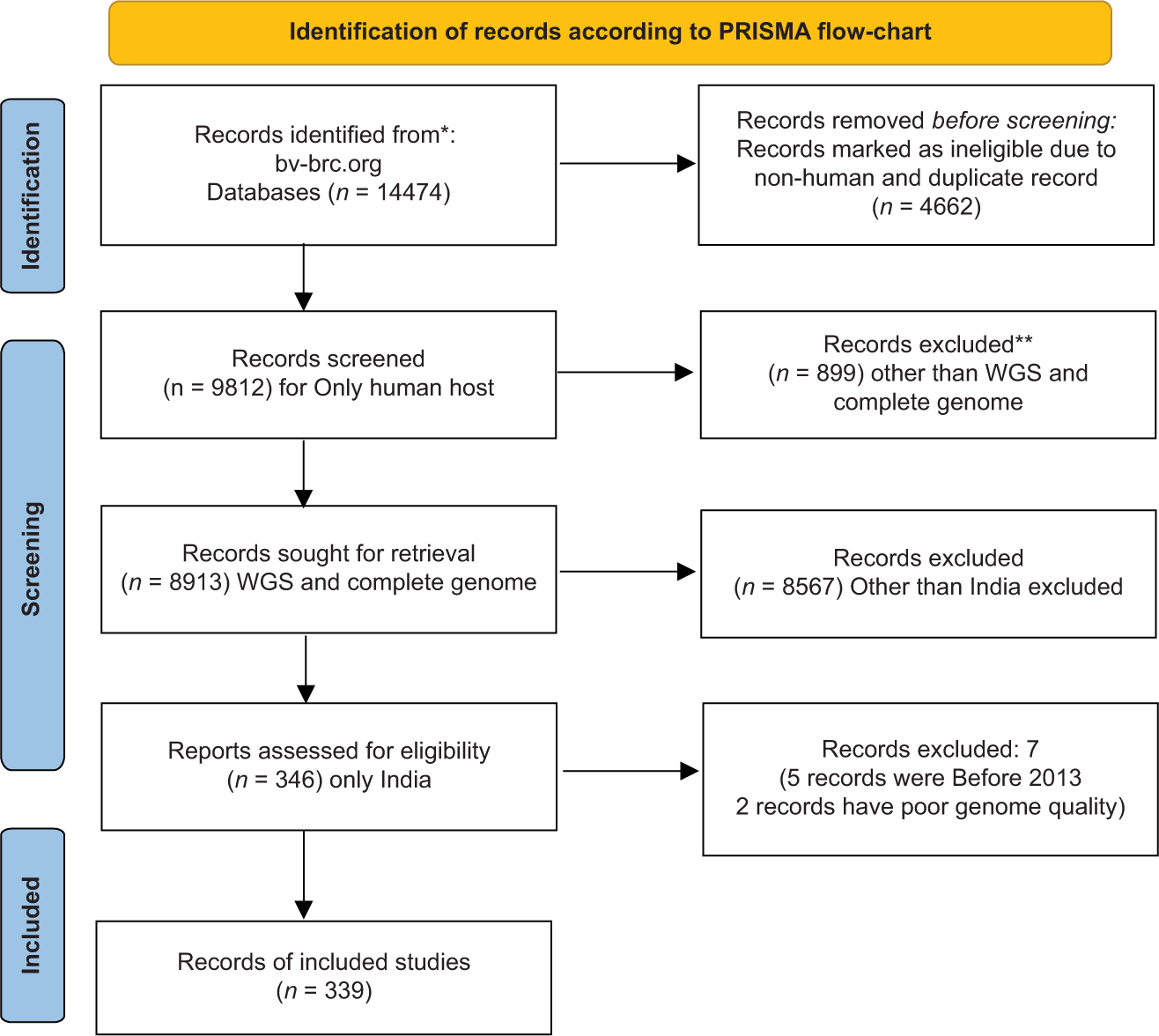
- Identification of records according to PRISMA guidelines.
Out of 339 collected samples of A. baumannii isolates, 122 (36%) were from blood, 102 (30%) from endotracheal aspirate (ETA), 9 (2.65%) from sputum, 3 (0.88%) pus, 87 (25.66%) from the respiratory specimen, 3 (0.88%) from bronchoalveolar lavage (BAL), and 13 (3.83%) from other sources for genome sequencing in India during 2013–2020. The highest samples were collected from blood (36%), following the ETA (30%) [Figure 2].
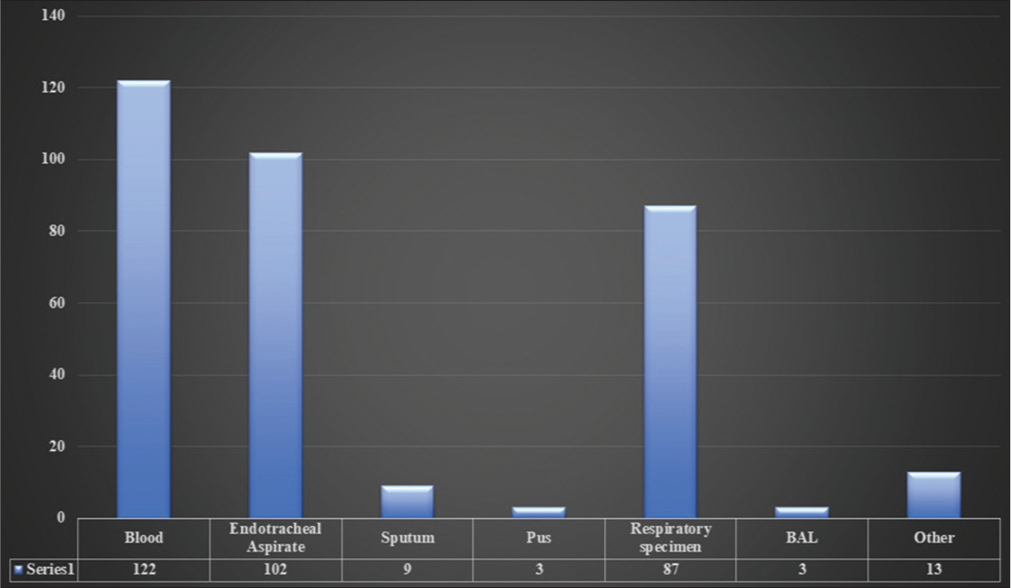
- Sample source.
[Table 1 and Figure 3] show the year-wise collection of the A. baumannii isolates.
| Collection Year | Total no. of isolates | Percent |
|---|---|---|
| 2013 | 2 | 0.6 |
| 2014 | 4 | 1.17 |
| 2015 | 8 | 2.35 |
| 2016 | 2 | 0.6 |
| 2017 | 11 | 3.24 |
| 2018 | 5 | 1.47 |
| 2019 | 218 | 64.30 |
| 2020 | 89 | 26.25 |
| Total | 339 |
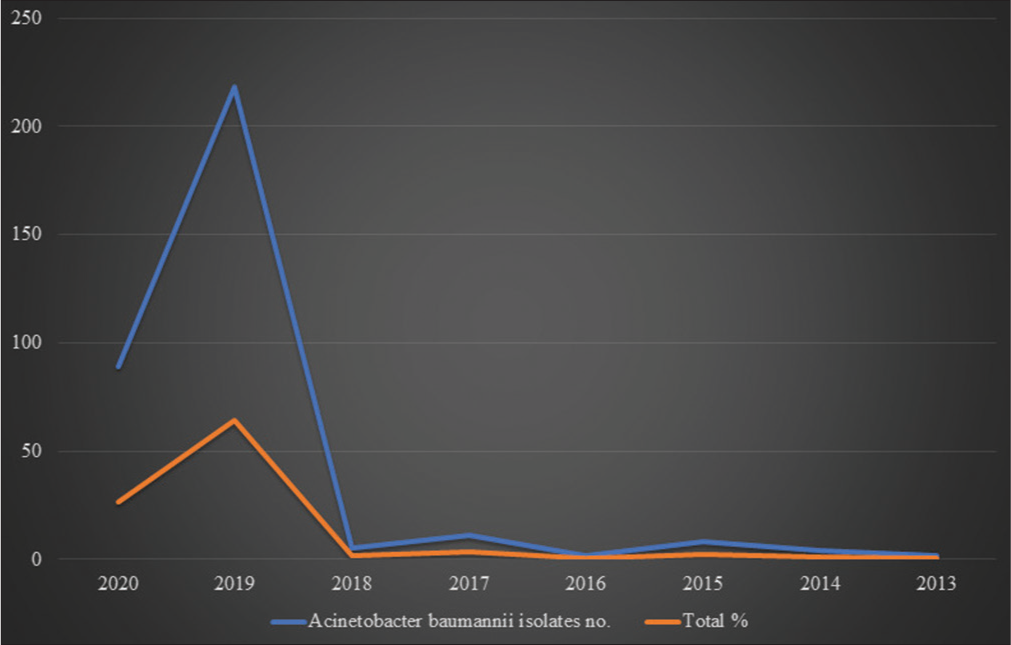
- Graphical representations of collected Acinetobacter baumannii isolates from 2013 to 2020.
In India during 2019, a maximum A. baumannii isolates (218) were collected, whereas, in 2013, only two isolates were collected for genome sequencing. Genome sequencing was done using various platforms, such as Illumina HiSec, Ion torrent, PacBio, and Oxford Nanopore MiniIon, and various assembly methods were used in these Bio projects such as SPAdes v. 5, SPAdes 3.14.1, Canu v.v 1.6, Canu v.v 1.7, UniCycler v.0.4.6, and UniCycler v.0.4.8. Typing was done via Multi Locus Sequence typing (MLST). The average genome size was 4,002,226 bp. The maximum genome size was 4,686,849bp in A. baumannii strain 2992, whereas the minimum genome size was 3,620,806bp in A. baumannii strain SP25. The average G+C% content was 38.95%. Among the 339 A. baumannii isolates, a maximum of 189 (55.75%) strains caused pneumonia, whereas 113 (33.33%) strains were involved in bacteremia.
PATRIC identified a total of 17540 antibiotic-resistant genes, CARD identified 10265 resistant genes, and through NDARO, 5112 were identified (as mentioned by bv-brc.org website). [Figure 4] depicts the antimicrobial resistance profile of all the genomes identified by PATRIC software. 157 A. baumannii strains were susceptible to various antimicrobials, 262 were resistant, and 81 were intermediate. Carbapenems seemed effective, but resistance against them was higher.
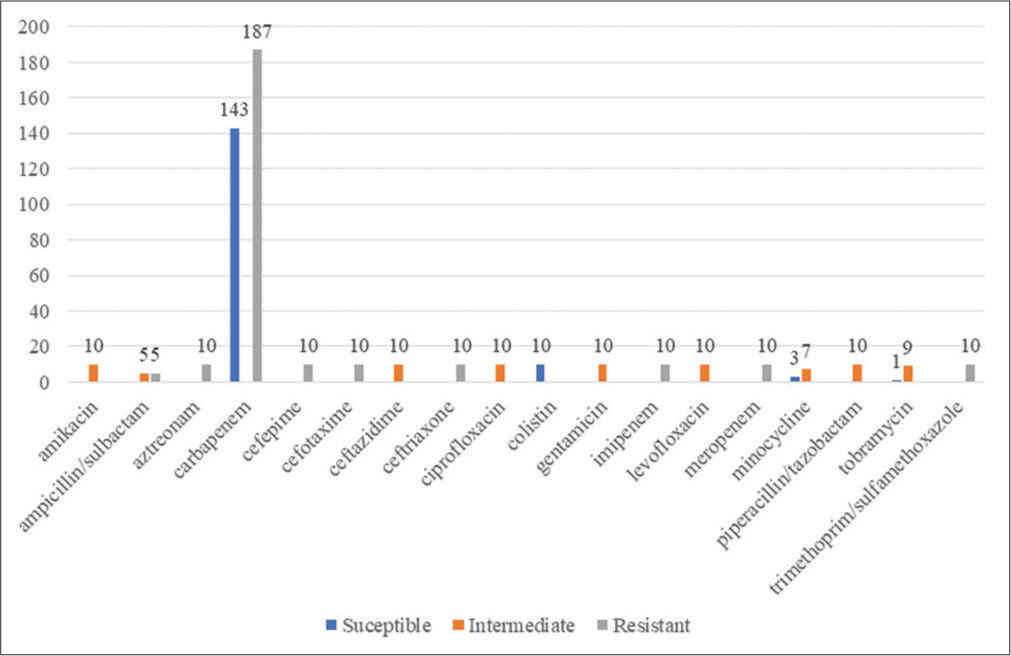
- Antimicrobial resistance profile of the various Acinetobacter baumannii isolates’ genomes.
[Figure 5] shows the frequency of various bla-OXA genes. Among all A. baumannii genomes, bla-OXA-23 had the highest frequency (314; 92.62%), followed by bla-OXA-66 (241; 71.09%) in India. Both enzymes were found in the A. baumannii genome along with other Carbapenemases such as NDM-1, TEM-1, ADC2, PER-7, PER-1, and AmpC genes and are highly involved in bacteremia and pneumonia. OXA-23 was found in almost all A. baumannii isolates (92.62%) except the following listed strains in [Table 2]. Out of 339 A. baumannii isolates, in 11 isolates, we did not find any resistant gene in their genome.
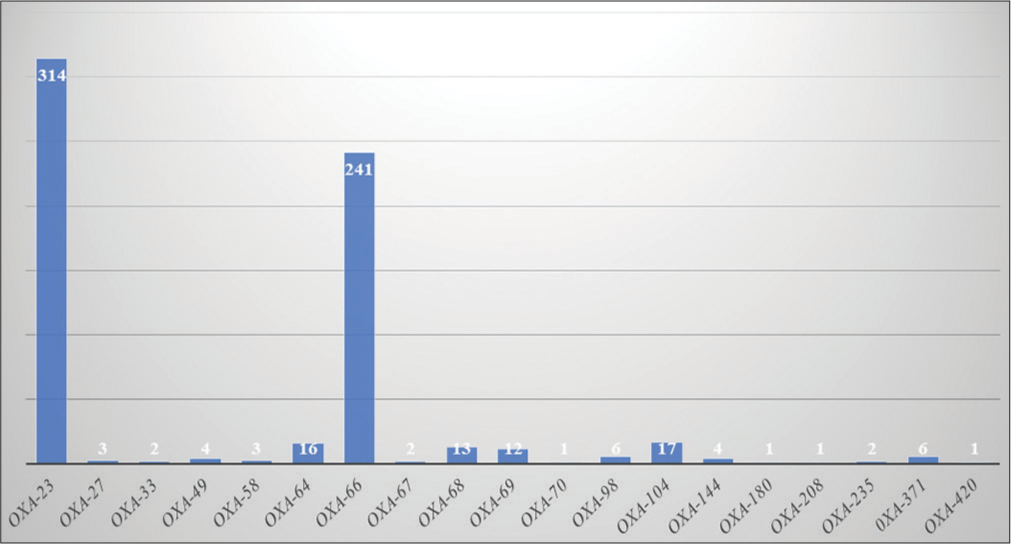
- Frequency of Oxacillinase encoding genes.
| S. No. |
A. baumanniistrain | Resistant genes |
|---|---|---|
| 1. | A. baumannii strain SP1917 | AmpC |
| 2. | A. baumannii strain SP25 | OXA-208, ADC2 |
| 3. | A. baumannii strain SP2128 | OXA-67, ADC2 |
| 4. | A. baumannii strain SP2210 | OXA-67, ADC2 |
| 5. | A. baumannii strain VB24319 | OXA-371, NDM-1, ADC2 |
| 6. | A. baumannii strain VB280820 |
OXA-58, OXA-64, CARB-1, NDM-1 |
| 7. | A. baumannii strain VB280821 |
OXA-58, OXA-64, CARB-1, NDM-1 |
| 8. | A. baumannii strain B8342 | OXA-33, AmpC |
| 9. | A. baumannii strain B11911 | PER-1, AmpC |
| 10. | A. baumannii strain B8300 | OXA-33, ADC2 |
| 11. | A. baumannii strain BA20352 | OXA-98, ADC2 |
| 12. | A. baumannii strain BA20475 | OXA-98, ADC2 |
| 13. | A. baumannii strain PM4229 | PER-7, OXA-68, ADC2 |
| 14. | A. baumannii strain PM194229 | PER-7, OXA-68, ADC2 |
A. baumannii: Acinetobacter baumannii
Among all the other genes, ADC2 was found more frequently (323) following NDM-1, in 281 strains. OXA-70 was found only in A. baumannii strain BP8282, OXA-180 in A. baumannii strain KSK sensitive, OXA-208 in A. baumannii strain SP25, and OXA-420 in A. baumannii strain SP4955.
[Table 3] shows that total of 12 oxacillinase genes blaOXA-64, blaOXA-66, blaOXA-67, blaOXA-68, blaOXA-69, blaOXA-70, blaOXA-98, blaOXA-104, blaOXA-144, bla-OXA180, blaOXA-208, and blaOXA-371 are similar to blaOXA-51 which is chromosomally encoded, unique for only A. baumannii and provides intrinsic resistance to this pathogen. All OXA-51 like carbapenem hydrolyzing enzymes have a chain of 274 amino acids and conserved domain for class D beta-lactamase (YbxI superfamily) at the position from 43 to 274 except OXA-69, OXA-208, and OXA-371 which have conserved domain interval for class D beta-lactamase at the position from 52 to 274.
| S. No. |
Oxacillinase genes |
Similarity | Conserved domain interval (YbxI superfamily) |
|---|---|---|---|
| 1. | bla-OXA 23 | bla-OXA 23 | 30-273 |
| 2. | bla-OXA 27 | bla-OXA 23 | 30-273 |
| 3. | bla-OXA 33 | bla-OXA 24 | 50-275 |
| 4. | bla-OXA 49 | bla-OXA 23 | 30-274 |
| 5. | bla-OXA 58 | bla-OXA 58 | 40-276 |
| 6. | bla-OXA 64 | bla-OXA 51 | 43-274 |
| 7. | bla-OXA 66 | bla-OXA 51 | 43-274 |
| 8. | bla-OXA 67 | bla-OXA 51 | 43-274 |
| 9. | bla-OXA 68 | bla-OXA 51 | 43-274 |
| 10. | bla-OXA 69 | bla-OXA 51 | 52-274 |
| 11 | bla-OXA 70 | bla-OXA 51 | 43-274 |
| 12. | bla-OXA 98 | bla-OXA 51 | 43-274 |
| 13. | bla-OXA 104 | bla-OXA 51 | 43-274 |
| 14 | bla-OXA 144 | bla-OXA 51 | 43-274 |
| 15. | bla-OXA 180 | bla-OXA 51 | 43-274 |
| 16. | bla-OXA 208 | bla-OXA 51 | 52-274 |
| 17. | bla-OXA 235 | bla-OXA 134 | 39-273 |
| 18. | bla-OXA 371 | bla-OXA 51 | 52-274 |
| 19. | bla-OXA 420 | bla-OXA 58 | 40-276 |
blaOXA-23, blaOXA-27, blaOXA-33, and blaOXA-58 are plasmid-mediated genes. Through horizontal gene transfer, transposons, and integron gene cassettes, they can cause a clonal outbreak of MDR A. baumannii in the ICU. Nosocomial infections caused by them are very difficult to treat, resulting in higher morbidity, mortality, and increased hospital stay of patients that cause a burden on both the patients and the hospitals.
blaOXA-23 and bla-OXA-27 share a similarity with each other. Both have 273 amino acids long chains in which the conserved domain for class D beta-lactamase is found at the position 30-273.
blaOXA-33 comes under the OXA-24 family. It has 275 long amino acid chains and the conserved domain for beta-lactamase lies between 50–275.
blaOXA-58 shares similarities with blaOXA-420 with 280 long amino acid residues chain and conserved domain for class D beta-lactamase found at the position between 40–276.
bla-OXA-235 comes under the OXA-134 family, and its length of amino acids chain is 276. The conserved domain for this enzyme is found at the positions 39–273.
[Table 4] depicts identities, positive values, and the gap among all the OXA-51 variants enzymes’ FASTA sequences, found during protein BLAST.
| S. No. | OXA-51 like family | Length | Identities | Identities % | Positives | Positives % | Gap |
|---|---|---|---|---|---|---|---|
| 1. | OXA-64 | 274 | 271 | 98.90 | 271 | 98.90 | - |
| 2. | OXA-66 | 274 | 268 | 97.81 | 270 | 98.54 | - |
| 3. | OXA-67 | 274 | 268 | 97.81 | 269 | 98.17 | - |
| 4. | OXA-68 | 274 | 266 | 97 | 270 | 98.54 | - |
| 5. | OXA-69 | 274 | 266 | 97 | 270 | 98.54 | - |
| 6. | OXA-70 | 274 | 272 | 99.27 | 273 | 99.63 | - |
| 7. | OXA-98 | 274 | 267 | 97.44 | 270 | 98.54 | - |
| 8. | OXA-104 | 274 | 268 | 97.81 | 270 | 98.54 | - |
| 9. | OXA-144 | 274 | 265 | 96.71 | 270 | 98.54 | - |
| 10. | OXA-180 | 274 | 270 | 98.54 | 271 | 98.90 | - |
| 11. | OXA-208 | 274 | 267 | 97.44 | 269 | 98.17 | - |
| 12. | OXA-371 | 274 | 265 | 96.71 | 270 | 98.54 | - |
BLAST: Basic local alignment search tool
[Figure 6] shows the phylogenetic tree of class D oxacillinase enzymes in which evolutionary history was inferred using the Neighbor-Joining method. The optimal tree is shown (next to the branches). The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The proportion of sites where at least one unambiguous base is present in at least one sequence for each descendent clade is shown next to each internal node in the tree. This analysis involved 22 amino acid sequences. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 284 positions in the final dataset.

- Phylogenetic tree of Oxacillinase enzymes.
By protein BLAST with OXA-51, OXA-23, OXA-24, OXA-58, and OXA-134 sequence, we have found substitution and mismatch of amino acids at the following positions.
We did not find a substitution of amino acids between OXA-51 and OXA-64, and by analyzing the evolutionary phylogenetic tree, we have found that both arose from the same internal node. OXA-64 is more similar to OXA-51 among other variants. The mismatch was found at the position no. 5, where alanine replaced threonine; at the position 38, glycine replaced alanine; at the position no. 48, alanine replaced valine.
OXA-69 and OXA-371 originated from the same internal node, and both are more closely related than OXA-66. Both OXA-69 and OXA-371 have glutamic acid instead of glutamine at the position no. 107, and asparagine at the position no. 117 instead of aspartic acid, asparagine at the position no. 225 instead of aspartic acid. OXA-371 differs from OXA-69 as it has aspartic acid at the position no. 36 instead of glutamic acid and lysine at position no. 237 instead of glutamine.
OXA-69 and OXA-371 have mismatched amino acids at the position no. 5, where alanine was found instead of threonine: at the position 48, alanine replaced valine, and at the position 57, histidine was located instead of glutamine. At position no. 194 in OXA-69, glutamine replaced proline, whereas in OXA-371, proline replaced glutamine.
OXA-66 has a lysine at the position no. 107 instead of glutamine, and at the position no. 125, asparagine instead of aspartic acid: thus, it is different from OXA-69 and OXA-371. The OXA-66 FASTA sequence was mismatched at the positions 5, 48, and 194, the same as in OXA-69, but at the position no. 36 in OXA-66, valine replaced glutamic acid.
OXA-67 arises as a separate internal branch. At the position no. 101, isoleucine substituted valine. The mismatch was found at the position 5, where alanine replaced threonine; at the position no. 12 threonine replaced alanine; at the position no. 31 threonine replaced alanine; at the position no. 48 alanine replaced valine, and at the position no.194 leucine replaced proline.
OXA-68 and OXA-144 are closely related. Both have asparagine at the positions no. 117 and 225 instead of aspartic acid and serine at the position 24 instead of threonine, but OXA-144 also has a substitution of amino acids at the position 195, where glutamic acid replaced lysine; and at the position 203 isoleucine replaced methionine. Mismatches were found in both OXA-68 and OXA-144 enzymes at the same positions, such as the position no. 5, where alanine was found instead of threonine; at the position 48, alanine instead of valine; at the position 146 asparagine instead of lysine, and at the position 194 glutamine was found instead of proline. However, in OXA-68, at the position no. 195 glutamic acid has replaced lysine.
OXA-98 arises as a separate branch, and it is precisely similar to OXA-68, where the same amino acids substituted others at the same positions no. 24,117 and 225. The mismatched position was also identical at the positions 5, 48, 146, and 194, but the only difference is at the position no. 195 in OXA-68 where a mismatch was found, but not in the OXA-98.
OXA-70 and OXA-208 share the same internal node and are variants of OXA-51, like other OXA-51 like enzymes. In the OXA-70, at the position 39, glutamic acid substituted alanine; whereas at the position 108, glutamine replaced glutamic acid. OXA-70 was found only in A. baumannii strain BP8282, whereas OXA-208 was found only in A. baumannii strain SP25. Two substitution positions were found in the OXA-208: at the position 36, aspartic acid substituted glutamic acid; at the position 105, asparagine was located instead of aspartic acid. There are five mismatches: at the positions 5, 48, 57, and 194, same as OXA-69 and OXA-371, and at the position 146, asparagine replaced lysine as in OXA-68 and OXA-144.
OXA-104 has a separate branch. Substituted amino acids were found at the position 107, where lysine replaced glutamine, and asparagine replaced aspartic acid at the position 117. Mismatch amino acid positions were found same as OXA-144.
OXA-180 has a distinct branch where substitution was found at the position 124 only (isoleucine substituted methionine). Mismatches were found at the position 5, where alanine replaced threonine; at the position no.48, alanine replaced valine; and at the position no.194, glutamine replaced glutamic acid.
OXA-23 and OXA-27 are 99.26% similar with 273 long amino acid chain. Substitution of amino acids was not found between them. Mismatched amino acids were found at the position no. 95, where alanine has replaced threonine, and at the position no. 247, lysine has replaced asparagine.
OXA-33 and OXA-24 both have 275 long amino acid chains and are 99% identical, and at the position 238, glutamine replaced arginine. OXA-33 is seen only in A. baumannii strain B8300 with ADC2 gene and in A. baumannii strain B8342 with AmpC gene, whereas OXA-24 is seen in Acinetobacter calcoaceticus and A. baumannii complex. We did not find OXA-24 in our 339 comparative genome group of A. baumannii isolates between 2013 and 2020.
OXA-58 and OXA-420 are 99.64% identical and aroused from the same internal node with 280 long amino acid chains. Both differ due to mutation at the position no. 256, whereas in OXA-420 aspartic acid replaced alanine. Substitution of amino acids was not found between them.
OXA-235 is similar to OXA-134 with 276 long amino acids chain and is 99% identical to it. No substitution was found between them; only a mismatch was found at the position no. 109 in OXA-235, where alanine has replaced valine.
OXA-49 is 99% identical to OXA-23, but OXA-49 has 274 amino acids long chain instead of 273, and only one amino acid substitution was found at the position 148 (glutamic acid has replaced lysine). While pair-wise alignment with OXA-23, one gape was found at the position no.120 in OXA-49, and one extra copy of alanine was found at this the position.
DISCUSSION
A. baumannii is an opportunistic non-fermenting Gram-negative bacillus that is generally found in the hospital environment and, by producing biofilm, acquiring various resistance genes, or overexpressing these genes, can survive in harsh conditions and typically infect critically ill patients who are on ventilator support or in an immunocompromised state.
Oxacillinase is classified as class D in Ambler’s classification of beta-lactamases. The first OXA enzymes were penicillinases that could hydrolyze oxacillin and penicillin and impart resistance. This class has a weak hydrolysis activity against carbapenems and generally transfers through plasmids.[12] Despite their modest activity, these enzymes can drastically increase the isolate’s carbapenem minimum inhibitory concentration (MIC), leading to the development of resistance. The acquisition of OXA enzymes that can bestow carbapenem resistance on A. baumannii has made this resistance a critical challenge that threatens to impair the clinical efficacy of the carbapenems and are the main culprit of the clonal outbreak in the ICU that is very difficult to treat and increases morbidity and mortality.[13]
In our systematic review, we have found that in India, the prevalence of OXA-23 (92.62%), following OXA-66 (71.09%) is very high, and even all over the world, the prevalence of both resistance genes is high. Resistance to carbapenems was identified as the most common.
Various researchers, such as Veeraraghavan et al. (2019) and Kumar et al. (2019) in their research study, found a high prevalence of OXA-23 (97%) in India.[14,15] Cortivo et al. in 2015, investigated OXA-genes in A. baumannii and found a higher prevalence of OXA-23 (87%) in Brazil.[16] Yang et al., 2018 demonstrated that 94.17% of A. baumannii strains possessed blaOXA-23 genes and were non-susceptible to sulbactam.[17] Palmieri et al., 2020 stated that the most often identified, bla-OXA-23, appeared to be associated with carbapenem resistance, and the insertion sequence’s (ISAba-1) proximity to the allele likely boosted the production of this enzyme.[18,19] Afshar et al., 2022 demonstrated the significant role of OXA-23 against carbapenems, amikacin, and ampicillin/sulbactam.[20] OXA-51 is chromosomally encoded and a biomarker for identifying A. baumannii among other Acinetobacter spp. It provides intrinsic resistance to carbapenems, especially against imipenem. Sharma et al., 2020 found in their research study that among 150 isolates of A. baumannii, all the isolates possessed the OXA-51 gene, and 98% of the isolates had the OXA-23 gene.[21] According to Chandra et al., 2022, India has a prevalence of both the resistance genes blaOXA and bla-NDM. India is witnessing a surge in carbapenem-resistant A. baumannii International clone-2 (IC-2). Antibiotic resistance (AMR) hikes in healthcare expenses have a negative impact on 80% of Indians without insurance. The administration of tigecycline and polymyxins as the last alternative has worsened the prognosis due to the high dependence on carbapenems.[22]
A. baumannii has an overabundance of genomic diversity, allowing it to spread AMR genes that are frequently linked to mobile genetic elements. Analyzing the genetic surroundings of resistance genes often yields insightful data on the emergence, mutation, and dissemination of resistance. In order to improve infection control procedures and create antimicrobial stewardship strategies, it is essential to provide an appropriate, simple, and affordable diagnostic method to discover Carbapenemases production by A. baumannii strains at health facilities.
CONCLUSION
Our findings indicated that a high percentage of A. baumannii strains that produce oxacillinases exist in India, emphasizing the necessity for indigenous molecular surveillance to assist effective management and preventative initiatives. Statistics from nations lacking routine surveillance are essential to comprehend and prevent the emergence of carbapenem-resistant A. baumannii strains, which are now epidemic. Developing suitable infection control measures and more effective treatment approaches depends on our current understanding of A. baumannii strains outbreaks. Comparative genomics and next-generation sequencing techniques will offer tremendous potential for tracking and regulating the spread of this dangerous bacterium.
Acknowledgment
The authors thank Prof. R.G. Saini, (The Chairperson of Centre for Interdisciplinary Biomedical Research and Registrar, Adesh University) for his motivation and support.
Author’s Contribution
Authors Manita Paneri and Vipul D. Yagnik# have contributed equally to the article.
Declaration of patient consent
Patient’s consent not required as there are no patients in this study.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Available from: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed [Last accessed on 2023 Jan 15]
- Clinical and genomic analysis of virulence-related genes in bloodstream infections caused by Acinetobacter baumannii. Virulence. 2022;13:1920-7.
- [CrossRef] [PubMed] [Google Scholar]
- Carbapenem use in critically ill patients. Curr Opin Infect Dis. 2020;33:86-91.
- [CrossRef] [PubMed] [Google Scholar]
- Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021;10:373.
- [CrossRef] [PubMed] [Google Scholar]
- The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int J Surg. 2021;88:105906.
- [CrossRef] [PubMed] [Google Scholar]
- Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51:D678-89.
- [CrossRef] [PubMed] [Google Scholar]
- Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535-42.
- [CrossRef] [PubMed] [Google Scholar]
- Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101-9.
- [CrossRef] [PubMed] [Google Scholar]
- The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-25.
- [Google Scholar]
- Evolutionary divergence and convergence in proteins In: Bryson V, Vogel HJ, eds. Evolving Genes and Proteins. New York: Academic Press; 1965. p. :97-166.
- [CrossRef] [Google Scholar]
- MEGA 11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022-7.
- [CrossRef] [PubMed] [Google Scholar]
- Rapid detection of OXA-23-like, OXA-24-like, and OXA-58-like carbapenemases from Acinetobacter species by real-time PCR. J Hosp Infect. 2020;105:741-6.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization & epidemiology of carbapenem-resistant Acinetobacter baumannii collected across India. Indian J Med Res. 2019;149:240-6.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates reveals the emergence of blaOXA-23 and blaNDM-1 encoding international clones in India. Infect Genet Evol. 2019;75:103986.
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial resistance profiles and oxacillinase genes in carbapenem-resistant Acinetobacter baumannii isolated from hospitalized patients in Santa Catarina, Brazil. Rev Soc Bras Med Trop. 2015;48:699-705.
- [CrossRef] [PubMed] [Google Scholar]
- OXA-23 Is a prevalent mechanism contributing to sulbactam resistance in diverse Acinetobacter baumannii clinical strains. Antimicrob Agents Chemother. 2018;63:e01676-18.
- [CrossRef] [PubMed] [Google Scholar]
- Abundance of colistin-resistant, OXA-23-and ArmA-producing Acinetobacter baumannii belonging to international clone 2 in greece. Front Microbiol. 2020;11:668.
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of OXA-Type β-lactamase genes among carbapenem-resistant Acinetobacter baumannii clinical isolates in Thailand. Antibiotics (Basel). 2020;9:864.
- [CrossRef] [PubMed] [Google Scholar]
- Molecular analysis of oxacillinase genes and identification of drug resistance pattern in MDR strains of Acinetobacter baumannii isolated from burn wound samples in Kermanshah, Iran. J Clin Diagn Res. 2022;16:DC14-8.
- [CrossRef] [Google Scholar]
- Distribution of carbapenemase genes in clinical isolates of Acinetobacter baumannii & a comparison of MALDI-TOF mass spectrometry-based detection of carbapenamase production with other phenotypic methods. Indian J Med Res. 2020;151:585-91.
- [CrossRef] [PubMed] [Google Scholar]
- Multidrug-resistant Acinetobacter baumannii infections: Looming threat in the Indian clinical setting. Expert Rev Anti Infect Ther. 2022;20:721-32.
- [CrossRef] [PubMed] [Google Scholar]







